Just noticed this while working and debugging covid5ync app. Don't know if that may have sense from a biotechnology view... still, it worth to be mentioned:
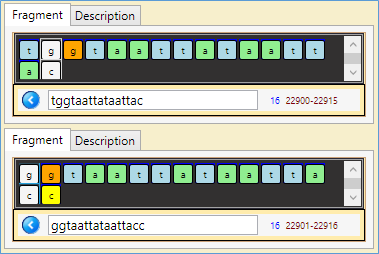
- A fragment (15 nucleotides) of the RNA sequence looks to be a 'palindrome' -:) of itself… Position 22901 -22916
Note: the top part of the figures below reads sequence's nucleotides in the direction 5'->3', the lower should be read in the reverse direction 3'->5' (which is the complementary of the upper part (i.e. 'a' complementary to 't' and 'g' to 'c'))…
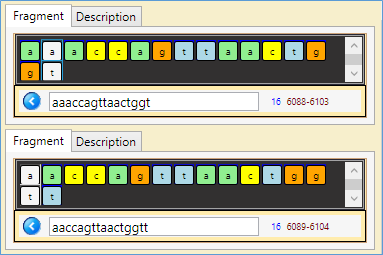
- A similar 'palindrome' fragment (15 nucleotides): 6089 - 6104
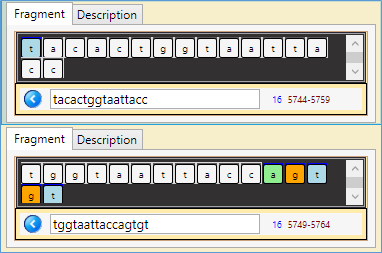
- Similar with a small shift: (5744 – 5764)
I continue posting about covid-5ync app (a helper app contribution in the fight against the latest coronavirus epidemic)
A major step now done: serializing application's information into xml files!
Hopefully this will allow transmitting research session work among members of our target community: biotechnology engineers.
I also downloaded (@NCBI site) a recent version of the virus RNA sequence (referred to as: MT163719 29903bp RNA linear VRL 10-MAR-2020). A daily work is done by the application to retrieve significant regions on the sequence. The updated version of the (xml) file is then uploaded to the project's web site which will allow gaining time in some tedious analysis tasks.
Serialization / deserialization
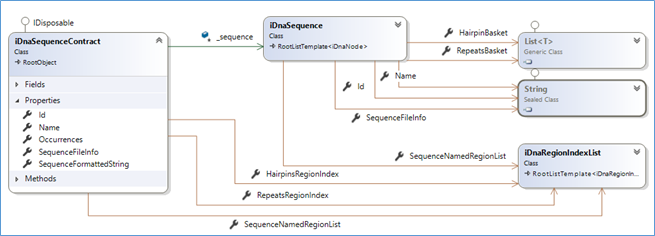
Reminder: Technically speaking, the sequence's data structures can be summarized as:
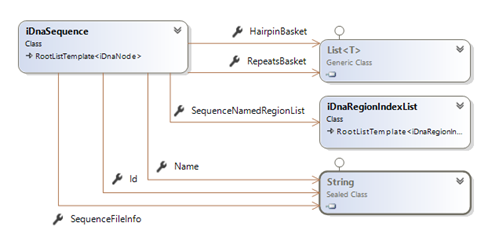
To serialize these information… (in regards of the urgency matters) I decided to simply use a DataContractSerializer (which, as I said in other posts, is not really the best solution for extensibility).
The encountered difficulty was on several aspects:
- Some information is redundant, notably for sub-sequences (collections of repeats and hairpins)
- The sequence object being itself a collection of nodes, the serializer did not really allow an easy way to handle its sub-objects. Here we fall into a known problem of cycling!
The solution used was:
- Transform sub-sequences to region lists (start/end indexes + name + occurrences) and deserialize back to sub-sequences
- Create a serialization-specialized object (using DataContract / DataMember attributes) and process the serialization through that object to avoid the 'cycling' errors.
The second difficulty was errors encountered on deserialization while locating and assigning various regions start/end indexes to the sequence's nodes.
Actually, the deserialization submits the sequence's node as a string to be parsed asynchrony. And as you may have guessed, the collection of sequence's nodes was being altered while locating the regions' nodes!
The solution:
- The Parse method raises Parse Complete event
- Subscribe to that event and proceed to these operations.
That is the short story… will keep you posted about more details later this week!
KEEP SAFE: wash your hands + do not touch your face + keep hope: Humanity will prevail!