A quick post to talk about the progress in this project with a few technical details.
First, a screen capture of what it looks like today (just a little better than before!):
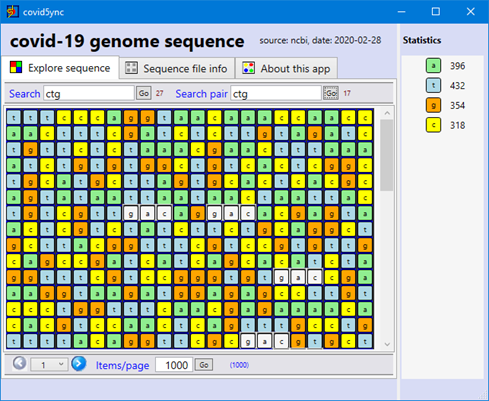
The first version of the application is already online: click-once installation. And Yes, we don't have money to have a certificate for the deployment… install only if you trust!... any suggestions about sponsors are also greatly welcome!
A few features implemented:
- Search for string and search for the 'complementary' of string.
- Paging
- Zoom-in / out on the sequence
Next steps:
- Search enhancement: visually locate occurrences
- Search for unique fragments according to user-provided settings
- Open file / save selection
Long-term:
Now, let us take a quick dive into the mechanics
The first class diagram (updated source code with such documentation is @github)
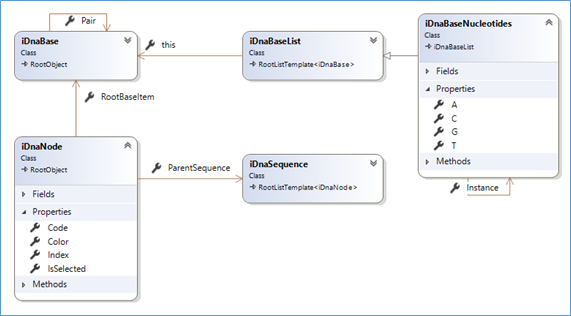
That seems quite basic(!), actually, as the reality of DNA, and that is one reason it is also fascinating!
- A sequence (iDnaSequence) is composed of 'nodes' (iDnaNode)
- A node is noted as a char. It refers to a 'base'.
- Commonly a set of 4 bases is used ('a', 't', 'g' and 'c'). There may be more, but we can easily handle this when needed.
- Each individual base has a complementary 'Pair'. A is pairable with T, C with G
- The 'standard' set of bases (iDnaBaseNucleotides) is a (singleton) list of the 4 bases. It sits as the main reference for nodes. It provides answers to important questions like: is 'c' a valid base?... what the complementary of 'X'?... what is the complementary sequence of the string "atgccca"? and so on.
Visual presentation: a start point
There are many ways to present a DNA sequence. To start with something, let us assign a color to each base. The user can later change this to obtain a view according to his or her work area. Technically speaking, we have some constraints:
- The number of nucleotides of a sequence can be important. For coronavirus, that is roughly 29000. We therefor need 'paging' to display and interact with a sequence.
- Using identification colors for nucleotides can also help to visually identify meaningful regions of the sequence on hand. For this to be useful, we need to implement zoom-in/out on the displayed sequence.
Paging
I simply used the solution exposed in a previous post about doc5ync.
Zoom-in / out
I found a good solution through a discussion on Stack Overflow
(credit: https://stackoverflow.com/users/967254/almulo).
- Find the scroll viewer of the ListView.
- Handle zoom events as required
var presenter = UiHelpers.FindChild<ScrollContentPresenter>(listItems, null);
var mouseWheelZoom = new MouseWheelZoom(presenter);
PreviewMouseWheel += mouseWheelZoom.Zoom;
|
Sample screen shots of a zoom-out / in

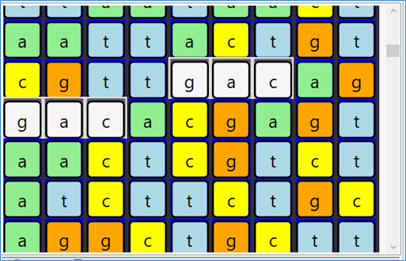
More details later in a future post.
Please send your remarks, suggestions and contributions on github.
Hope all that will be useful in some way… Time is running… Humanity will prevail